Loading a Map¶
Metabolic models annotated in the Systems Biology Markup Language (SBML) (Hucka et. al.) and BioPAX (Demir et. al.) formats are available in a number of different databases. These databases include, but are not limited to BiGG, MetExplore, Human Metabolic Atlas, Virtual Metabolic Human, BioModels, PathBank, and MetaCyc. These formats are used in the annotation of constraint-based metabolic reconstructions (O’Brien et. al.), a popular platforms for modeling metabolic networks. SAMMI accepts as input both SBML and BioPAX annotated models, and can also load metabolic networks annotated from KEGG. This section explains how to initially load these models.
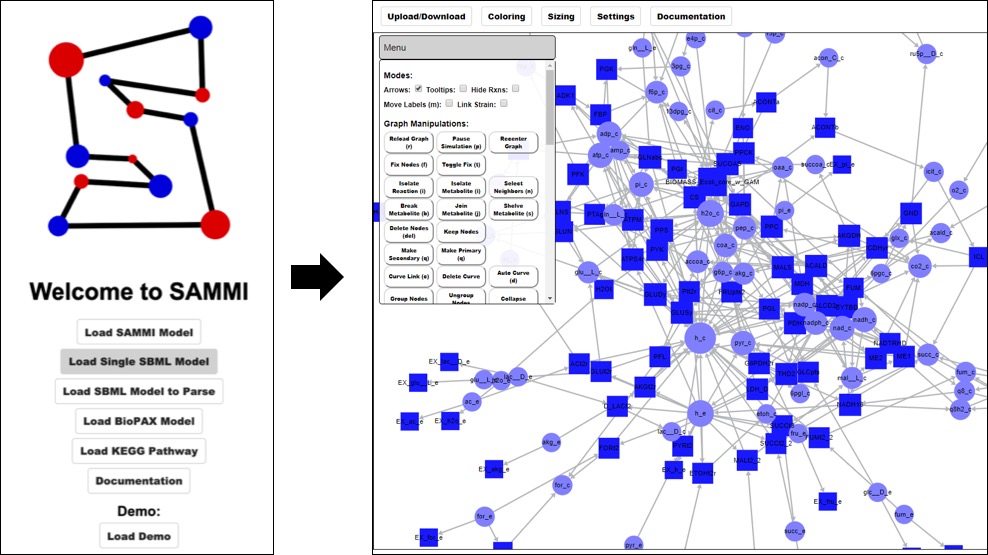
Load SBML Model¶
To load a single map for the entire reconstruction, Click on Load Single Model at the home screen and select the metabolic reconstruction file.
This option will load every reaction and metabolite in the reconstruction. This functionality is recommended only for relatively small models, as large networks will yield slow-rendering and convoluted maps. For large reconstructions, users can parse the model into multiple subgraphs during the upload process. In order to do so,click on Load Model to Parse at the home screen. This will bring you to a second menu where you can select whether to parse the graph based on model annotation or user-defined input. To generate a map based on model data, select either Reactions or Metabolites from the dropdown menu. This will populate a new dropdown menu with annotation categories from the model. Select a category and SAMMI will build a subgraph for each value under that data type. For instance, select Reactions then Subsystem to make a subgraph for each subsystem defined in the reconstruction, or select Metabolite then Compartment to make a map for each cellular compartment.
Parsing a model through reaction annotation can lead to the inclusion of metabolites in multiple subgraphs, but no more than one copy of each metabolite is initially loaded in each subgraph. If parsing a model through metabolite annotation, all reactions containing a metabolite with that annotation will be included in each subgraph. This can lead to the inclusion of metabolites with a different annotation in the subgraph. For instance, if parsing a reconstruction based on metabolite compartments, the mitochondria compartment will also include transport reactions through the mitochondrial membrane, which in turn will lead to the inclusion of cytosolic metabolites.
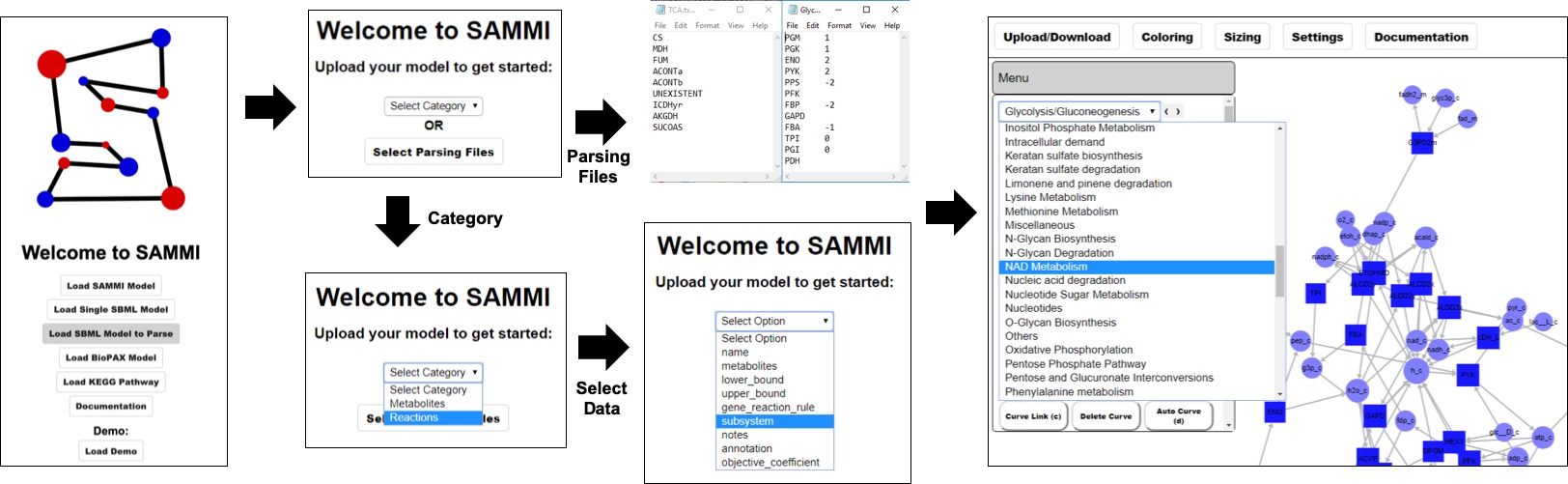
Models can be parsed in two different ways. Either using reaction or metabolite data annotated in the reconstruction (bottom) or using user-defined lists of reaction IDs.
The second option is to parse the model based on user-defined inputs. To use this option, users should define any number of tab-delimited text files with two columns: reaction IDs and optional associated numerical values. No header should be included. After defining the files and selecting Load Model to Parse, click on Select Parsing Files and select all the desired files. SAMMI will make a subgraph for each file selected. Here, reactions can be included in multiple subgraphs or no subgraph at all. Reactions with an associated value will retain that value as reaction color data. Reactions with no associate value will be included in the model but will have no associated data.
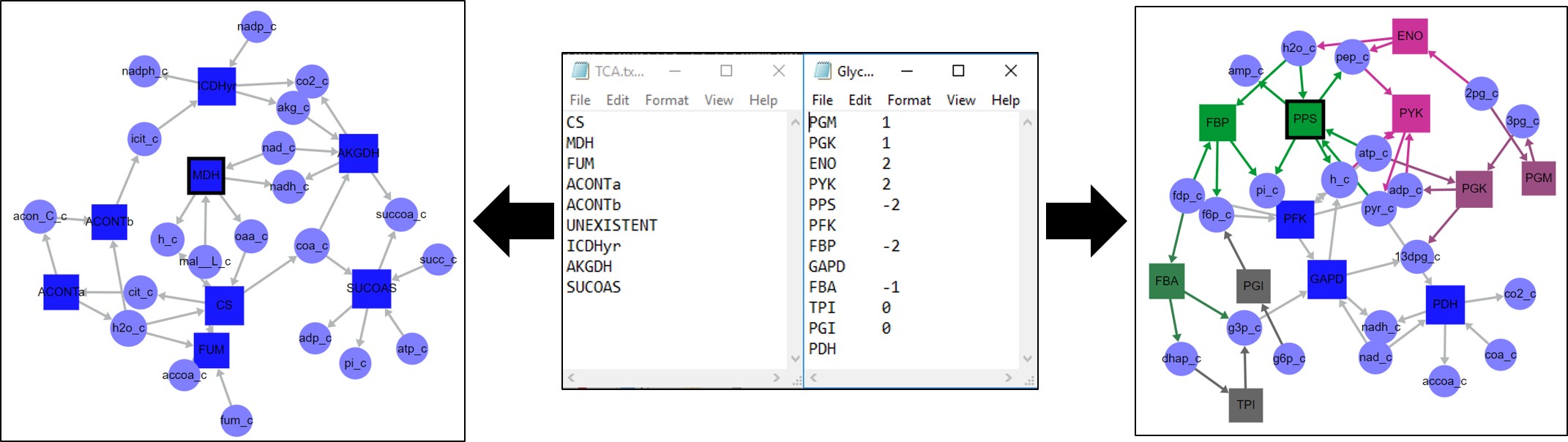
Two valid examples of model parsing files. The file on the left will load a map including only the reactions in that file, with no associated data. The line UNEXISTENT will be ignored since no such reaction is present in the model. The file on the right will load associated reaction data where those values are defined.
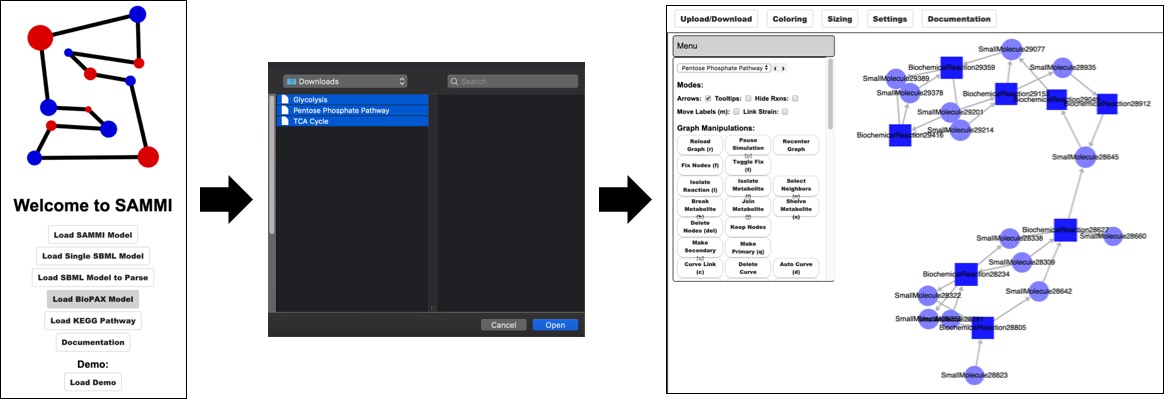
Load BioPAX Model¶
In oder to load BioPAX models select Load BioPAX Model in the SAMMI home screen. For this options users can select multiple files, each of which are loaded as a different subgraph. File names will be used to name each subgraph, although these names can be changed after models are loaded.
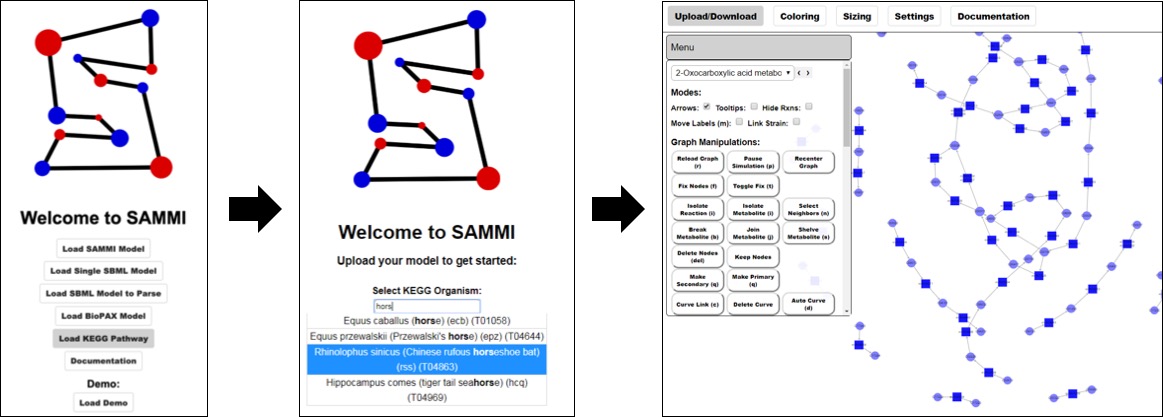
Load KEGG Pathway¶
The metabolic pathways for over 5,000 organisms have been made readily available in SAMMI. These pathways were obtained from KEGG in January of 2019 and contain associated metabolite and reaction annotations. To load a metabolic map from KEGG click on Load KEGG Pathway in the home screen. Next, type the name of the desired organism and matching names will be autocompleted. Organisms can be searched by binomial nomenclature, informal name, and organism KEGG IDs. Select the desired organism from the autocomplete list and hit enter to load the desired map.